Multicentre trials powered by XNAT for NCITA

XNAT is an advanced management system for research images. It uses multiple technologies to provide researchers with the tools they need to process data and view data. The ICR has been at the forefront of developing new capabilities for XNAT, in particular, the ICR-XNAT-OHIF viewer (see Doran et al. Tomography 8.1 (2022). Our most recent innovations on the viewer have been created in partnership with the National Cancer Imaging Translational Accelerator, funded by Cancer Research UK.
In 2018, XNAT technology was adopted by the National Cancer Imaging Translational Accelerator, a consortium of nine UK universities specialising in advanced diagnostic imaging, funded by Cancer Research UK. ICR staff manage and play key roles as members of the NCITA Repository Unit. This page lists the imaging studies powered by XNAT and supported by NCITA.
List of NCITA studies using XNAT through the NCITA Repository Unit
[1] NCITA Exemplar 3: The MUK NINE B trial – Establishing the environment for UK multi-centre clinical evaluation of whole-body (WB) MRI as a diagnostic and treatment response marker in multiple myeloma
FEATURED STUDY: NCITA EXEMPLAR 3, THE MUK NINE B TRIAL
Establishing the environment for UK multi-centre clinical evaluation of whole-body (WB) MRI as a diagnostic and treatment response marker in multiple myeloma
Project Leads: Prof Dow-Mu Koh, The Royal Marsden NHS Foundation Trust and Institute of Cancer Research (ICR), London; Dr Martin Kaiser, ICR, London (Chief Investigator) and Prof Christina Messiou, The Royal Marsden NHS Foundation Trust (Principal Investigator, Imaging Sub-study)
Whole-body magnetic resonance imaging (WB-MRI) is the most sensitive imaging test for diagnosis of myeloma bone marrow involvement and is recommended in the UK by NICE as the first line imaging test in patients with suspected myeloma.
The Myeloma Response Assessment and Diagnosis System (MY-RADS) guidelines promote the standardization of WB-MRI diagnosis and treatment response assessment of myeloma.
However, MY-RADS, which includes quantitative measurements, have not been prospectively validated or employed in studies guiding myeloma patient management. Furthermore, WB-MRI is currently limited to a few expert UK centres due to capacity limitations and technical and educational challenges.
The NCITA Exemplar 3 study (MUK Nine : OPTIMUM Treatment Protocol) ClinicalTrials.gov Identifier: NCT03188172 has expanded the availability of WB-MRI for multiple myeloma with the support of the NCITA QA/QC Unit and Image Repository Unit. Data for this study are hosted on the ICR/RMH Repository in its capacity as the ICR node of the NCITA Repository Unit. The XNAT Team facilitated the collection and anonymisation of volunteer and patient data from 13 institutions.

[2] NCITA Exemplar 4: Novel and multi-parametric (mp) MRI PROSTATE image repository for development of artificial intelligence automated reporting and multi-centre clinical trials. Project Lead: Prof Shonit Punwani (University College London). Data are stored on the UCL node of the NCITA Repository Unit
[3] NCITA Exemplar 5: OE-MRI for PAtients with Lung cancer (OPAL study). Project Leads: Prof James O’Connor (ICR & University of Manchester) and Prof Geoff Parker (UCL). Data are stored on the UCL node of the NCITA Repository Unit.
[4] NCITA Exemplar 7: The FIG trial – 18F-FDOPA PET imaging in GLIOMA: feasibility study for PET guided brain biopsy
FEATURED STUDY: NCITA EXEMPLAR 7, THE FIG TRIAL
18F-FDOPA PET imaging in GLIOMA: feasibility study for PET guided brain biopsy
Project Lead: Prof Geoff Higgins, University of Oxford
The radiotracer 3,4-dihydroxy-6-[ 18F] fluoro-L-phenylalanine ( 18F-FDOPA) has been shown to be a promising imaging agent for positron emission tomography (PET) imaging evaluation of brain tumours.
NCITA Exemplar 7 FIG study ( ClinicalTrials.gov Identifier: NCT04870580) is a multicentre Phase 1 study assessing the feasibility of performing 18F-FDOPA PET guided histopathology using standardised PET imaging protocols. Initially set up at the University of Oxford, and expanded other NCITA host institutions, including the University of Cambridge and Imperial College London.
The FIG study will also include assessment of the feasibility of oxygen-enhanced MRI (OE-MRI) for the detection of tumour hypoxia, in collaboration with The University of Manchester and UCL.
The ICR/RM Repository, in its capacity as the ICR node of the NCITA Repository Unit, hosts the data. Through a fruitful combination with the NCITA QA/QC Unit and MR Core Lab, advanced protocol verification software and a “containerised” processing workflow have been developed to allow data to be analysed efficiently and within tight timelines.
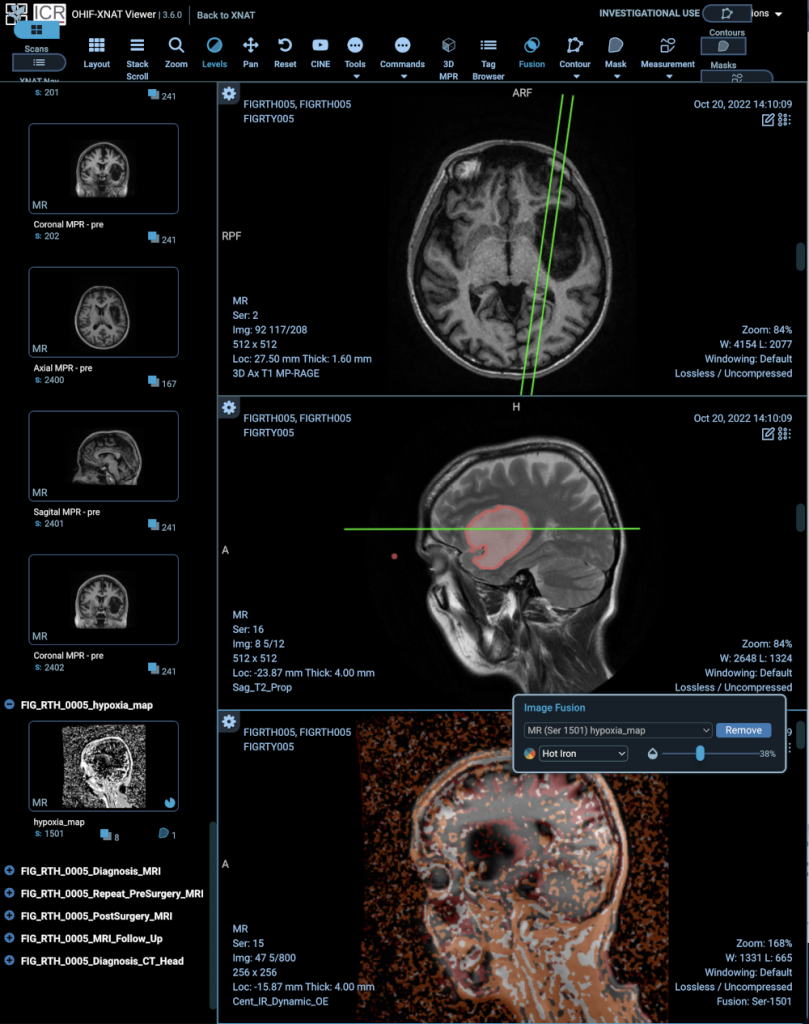
[5] NCITA adopted study EDIFICE: Early Detection of cancer recurrence In patients with glioblastoma using artiFicial IntelligenCE through imaging. Project Leads: Dr Stuart Currie (Leeds Teaching Hospitals NHS Trust), Dr Ali Gooya (University of Glasgow), Dr Patrick Hales (University College London), Dr Spencer Thomas (NPL Ltd). Data for this study are hosted on the ICR/RMH Repository in its capacity as the ICR node of the NCITA Repository Unit.
[6] NCITA adopted study AI and MR physics simulation to assess low-cost, low-field MRI as a cancer screening tool. Project Leads: Dr Matt Hall (NPL Ltd), Dr David Atkinson (University College London). Data for this study are hosted on the ICR/RMH Repository in its capacity as the ICR node of the NCITA Repository Unit.
[7] NCITA adopted study ARCADIAN: Atovaquone with Radical Chemoradiotherapy in locally advanced NSCLC. Project Lead: Prof Geoff Higgins (University of Oxford). Data for this study is hosted on the ICR/RMH Repository in its capacity as the ICR node of the NCITA Repository Unit.
[8] NCITA adopted study CRUK ACED Alliance: No data are stored by the Repository but Simon Doran sits on the ACED Information Governance and Data Protection board.
[9] NCITA adopted study ReIMAGINE: Consortium Director Prof Mark Emberton (University College London). ReIMAGINE Prostate Cancer Risk (NCT04060589) is a multi-centre, prospective, observational, longitudinal cohort study of men referred to secondary care with a suspicion of prostate cancer. The aim of the study is to develop a robust baseline risk stratification system for men at risk of prostate cancer. ReIMAGINE Prostate Cancer Screening (NCT04063566) is a single site study to assess the feasibility of prostate cancer screening using an invitation for a prostate MRI scan via GP practices. This feasibility study will assess the acceptability of an MRI as a prostate cancer screening assessment and assess the prevalence of MRI defined suspicious lesions and cancer in men across a spectrum of PSA results.Data are stored on the UCL node of the NCITA Repository Unit.
[10] NCITA adopted study LIBRA: Lung Nodule Imaging Biobank for Radiomics and AI Research (LIBRA) study. NCT04270799. Project Lead: Prof Richard Lee (Royal Marsden). Data for this study are hosted on the ICR/RMH Repository in its capacity as the ICR node of the NCITA Repository Unit.
[11] NCITA adopted study CLIMATE: Comparison of diagnostic accuracy of Luminal Index and Multi-parametric MRI for Accelerated deTEction of significant prostate cancer. Project Lead: Prof Shonit Punwani (UCL). Data are stored on the UCL node of the NCITA Repository Unit.
[12] NCITA adopted study LIMIT PCa: Luminal Index MRI Identification of Treatment critical Prostate Cancer. Project Lead: Prof Shonit Punwani (University College London). Data for this study are hosted on the ICR/RMH Repository in its capacity as the ICR node of the NCITA Repository Unit. The XNAT Team has developed an integrated workflow using the XNAT Container Service and XNAT Custom Forms, together with advanced 4-D image viewing features using the ICR-XNAT-OHIF viewer to streamline data processing, with the aim of a 30-minute turnaround time for data processing and reporting in a mobile MRI scanner.
[13] NCITA adopted study HERD: Multimodality Early Detection of Head and Neck Cancer Recurrence (NCT05097625). Project Lead: Prof Tony Ng (King’s College London). Data are stored on the UCL node of the NCITA Repository Unit.
[14] NCITA adopted study BRAID: Breast Screening – Risk Adaptive Imaging for Density (NCT04097366). Project Lead: Prof Fiona Gilbert (University of Cambridge). Data are stored on the Cambridge node of the NCITA Repository Unit.
[15] NCITA adopted study IIA-HGSOC: Integrated Image Analysis in High Grade Serous Ovarian Cancer. Project Lead: Prof Evis Sala (University of Cambridge, now Università Cattolica del Sacro Cuore). Data are stored on the Cambridge and ICR nodes of the NCITA Repository Unit.
[16] NCITA adopted study AI-COVID: AI-assisted diagnosis and prognostication in COVID-19. Project Lead: Prof Carola-Bibiane Schönlieb (University of Cambridge). Data are stored on the Cambridge node of the NCITA Repository Unit.
[17] NCITA adopted study LOOC: Lymphatic mapping of oropharyngeal cancer Study (NCT04498221). Project Lead: Prof Clare Schilling (University College London). Data for this study are hosted on the ICR/RMH Repository in its capacity as the ICR node of the NCITA Repository Unit and the XNAT Team has developed advanced image viewing facilities within XNAT to provide features for this trial.
[18] NCITA adopted study PANTHR-S: Precision medicine approaches for neoadjuvant therapy in high‐risk sarcoma patients
FEATURED STUDY: NCITA ADOPTED STUDY, PANTHR-S
Precision medicine approaches for neoadjuvant therapy in high‐risk sarcoma patients
Project leads: Prof Robin Jones (Royal Marsden), Dr Paul Huang (Institute of Canceer Research), Prof Christina Messiou (Royal Marsden and Institute of Cancer Research)
Development of a multifaceted data infrastructure to further understand sarcoma biology for patients with localized high-risk sarcoma undergoing perioperative management. Population group: patients with one of DDLPS, LMS, SS, vascular sarcomas or MPNST either advancing directly to surgery or receiving neoadjuvant chemotherapy and surgery. Types of data: radiology (CT and MRI), pathology, clinical (with link to genomic data).
The video shows how the ICR-XNAT-OHIF viewer is being used to develop a multimodal imaging experience that is able to bring all of these data together.

