Our technology: containers
XNAT is an advanced management system for research images. It uses multiple technologies to provide researchers with the tools they need to process data and view data. The ICR has been at the forefront of developing new capabilities for XNAT and were early adopters of XNAT’s Container service.
Goods for transportation come in all shapes and sizes. Containers allow shipping companies to take goods of any shape from door to door via land and see. Arbitrary consignments can be packed efficiently and standardised handling facilities are available anywhere.
Software containers are analogous to this. You can code your algorithm in any language. Your code is packaged with its own complete environment, so nothing in missing when you try to run it on another system. Every location where the software is run has its own standard handling facilities, so it doesn’t matter what model of computer you have or what the operating system is.

XNAT takes this concept a step further. Building on the Docker container-handling system, XNAT provides a Container Service that sits at the centre of a “triangle of providers”. The software developer – this could be a company or a postdoc in your lab – provides the algorithm as a container. The XNAT image archive supplies the data on which the algorithm runs. And the user provides the context in which the algorithm should run by choosing, either via a web browser or a program, which data need processing, what needs doing and when it needs to happen.

And here’s how it all comes together to implement the latest research from other imaging scientists around the world. The video below illustrates just how straightforward it is, as an XNAT user, to run advanced software that has been containerised. The ICR XNAT team just wraps everything up for you, so all you have to do is press one button! The video also showcases other developments that have been created and shared with the community by us, with funding from Cancer Research UK, via its NCITA consortium and in close collaboration with Flywheel. We have created a fully DICOM workflow, with outputs that could, potentially, flow back to the clinic. Our viewer integration features a highly sophisticated ROI contour panel that allows lazy loading of the structures created by Total Segmentator. The multiplanar view allows the contours to be displayed in any orientation, including oblique slices via a library that we have open-sourced.
But it’s not only about looking for external “products” and bringing them into our infrastructure. Containerisation also makes it possible to take new “home-grown” algorithms and deploy them for use in multi-centre research studies. These algorithms might include, as here an MRI-based segmentation (courtesy Antonio Candito and Matt Blackledge), or the calculation of so-called radiomics features, application of “defacing” algorithms to protect patient privacy, or even running the entire processing workflow for a clinical study.
Advances in technology are rapidly shifting the emphasis of data analysis away from the old paradigm in which the majority of a researcher’s effort would be to harmonise data from different centres, run a piece of software on each patient’s data and collate the results to a new paradigm whereby intellectual effort is focused, where where it should be, on design of the processing pipeline, leaving the container to do all the mundane tasks. This opens the possibility of re-running analyses with changed assumptions and re-producing the entire processing history a trial at the push of a button.

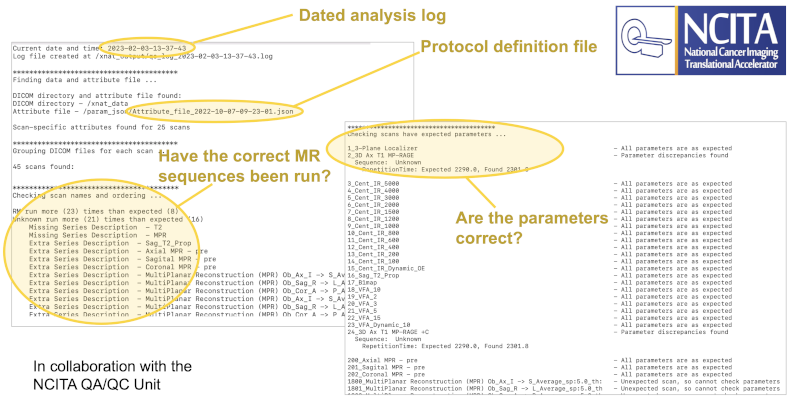
A vital part of running successful multicentre trials is the ability to ensure that incoming data from multiple sources is homogeneous in content. This is particularly important in MR studies, where images are acquired from different hospitals with a variety of scanner models from different vendors. The locally developed NCITA Protocol Checker (a collaboration between ICR and the University of Manchester) allows us to run a container that automatically verifies the content of the incoming images.
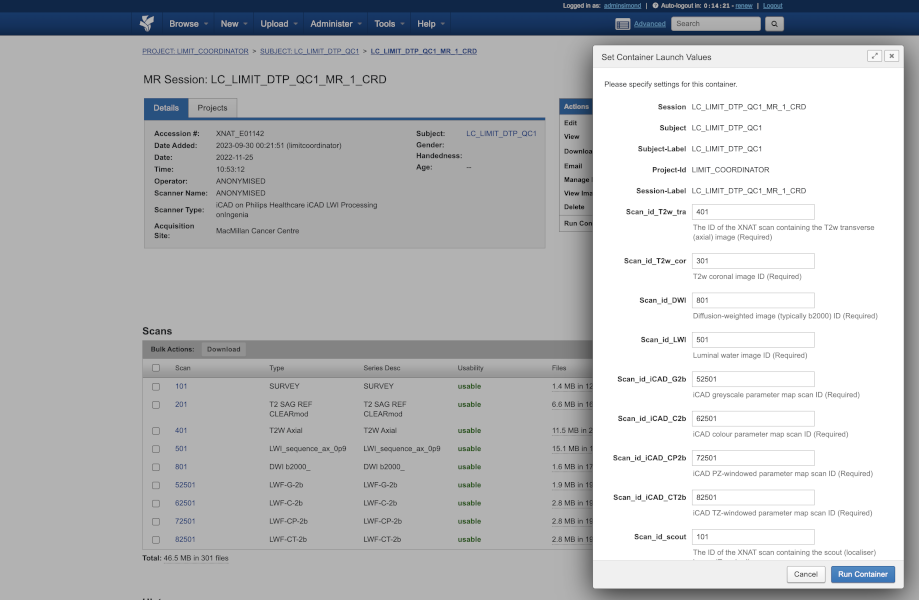
In the LIMIT trial, which is aimed at taking prostate cancer screening into the community, via a mobile MRI scanner, turnaround time from initial imaging to presenting the subject with their examination result will be crucial.
Containers allow us to orchestrate a complex process of sending data to a third-party processing pipeline hosted at ICR and then distributing different combinations of original and processed data to two different sets of radiologists to review results.