FAQs for Chief Investigators and Principal Investigators
How do I get started with XNAT?
Step 1: The study CI/PI should email the XNAT Project Lead, Simon Doran ([email protected]), explaining in broad terms what the research project consists of. As NCITA Repository Unit Manager, Simon is also a good first point of contact for new applications for adoption by NCITA of projects/multicentre studies for which the primary need is for data storage within the Repository Unit.
Step 2: Next, our project costing form should be filled in with as many details as you can regarding the type and quantity of data stored, and which of the Repository services you think you might need (e.g., data storage only, zero-footprint web-based image viewer, segmentation or other “containerised” analysis pipelines, deployment of AI algorithms, electronic case report forms, or bespoke coding). N.B. After you have done this, let a member of the XNAT Team know that you’ve filled it in, as Office 365 forms don’t automatically alert us.
Step 3: Depending on what you need the Simon will email you either a simple figure for your own costing purposes or a full statement of work and quotation. At this point, there may need to be some negotiation regarding exactly what will be delivered by the team and what the finally agreed cost will be. Once these details are locked down, we will record the details and agreement in an appropriate document for future reference.
Step 4: A representative of the team leading the study – this could be the CI/PI or Trial Coordinator – should then fill in our project setup form with all the information needed to create the XNAT projects that will be used to hold data for the study. This should include, as far as is known, the names and contact details of all the staff who will need userids on the XNAT platform in order to upload or access data. Note that, for a clinical trial/study, XNAT users with access to study data should all be included on the delegation log of the study.
Step 5: A meeting will be convened between the XNAT Team and the relevant CI or site PIs, at which the pseudonymisation and audit framework for the study will be discussed and an appropriate risk assessment signed off.
Step 6: For studies where data originate from sites outside the Royal Marsden or Institute of Cancer Research, the XNAT Team will contact a representative from each site to liaise regarding the technicalities of uploading data. We send an initial fact finding questionnaire to understand exactly what facilities are available. Based on this information, the XNAT Team will advise on a recommended upload method (see some of the possible options below). At this point it may be necessary to engage with IT or Information Security staff at the site to install relevant software and work through the upload process, resolving any difficulties that might be encountered.
Step 7: All XNAT users for the project need to receive appropriate training. For those not directly involved in data upload and anonymisation, it should be sufficient simply to read relevant parts of the Background Information and User SOPs for the ICR/RMH XNAT Repository and sign the declaration that they will abide by the code of conduct. For users involved in data upload, additional training is needed. This is currently organised by XNAT Team member Thesha Thavaraja ([email protected]).
Step 8: Once user training is complete and all signed documentation is in place, data upload can start. The XNAT Team will monitor initial data uploads to ensure that anonymisation is appropriate. Note that it is always the responsibility of the site staff to ensure that no identifiable data are sent to XNAT.
Step 9: The XNAT Team will then give access to the relevant members of the trial team (as specified on the project setup form) for them to analyse the incoming data. Should additional staff, not previously mentioned, need access to the data, then an access request form should be filled in for each user.
How do I make sure that my image data are anonymised correctly?
The XNAT Team has many years of experience in enabling users to upload their data safely to us. Depending on the facilities available at the sending institution, multiple routes are possible.

XNAT Desktop Client is currently the preferred method for uploading image data from outside the Institute of Cancer Research or Royal Marsden to our platforms. You can get a copy of this from the official XNAT download site. Once installed on your local computer, this app allows you to log into our XNAT server and choose the correct project, from where it “pulls” the appropriate anonymisation script. That script is designed by us (after a lengthy programme of research) to perform a standards-compliant, risk-assessed deidentification of your data. Deidentification happens on your own computer and thus inside your institution’s jurisdiction. The app lets you browse your local files to find the DICOM data to upload and then asks you to provide a pseudonymisation code. You are also required to visually inspect the data prior to sending, in order to ensure no unwanted patient-identifiable data are embedded in the image pixels themselves.
To use this method, you need to be able to install an application, usually on a hospital-administered computer. This may require permission from or the services of your local IT team. However, once installed you should require nothing more than regular internet access (technically, the HTTPS protocol on port 443). Normally, if you can see our website using a browser data upload should be straightforward.

DicomBrowser is a legacy piece of software that can nevertheless be extremely useful under certain circumstances. You can get a copy of this from the official XNAT download site. You can use the software in two ways: (i) to deidentify data and save the deidentified version locally; (ii) to deidentify the data and upload them directly to our platform. You will need an appropriate “anonymisation script” for your particular project and you can get this from the XNAT Team by emailing one of us. Method (i) above might be followed by upload using XNAT’s compressed uploader tool and this workflow is illustrated in this document.
To use DicomBrowser, you need to be able to install an application, usually on a hospital-administered computer. This may require permission from or the services of your local IT team. DicomBrowser is written in the Java programming language and this must be installed as well. If you want to use the software to send data to us directly using DicomBrowser, you may also need to liaise with your IT team regarding any local firewalls that may be in place.
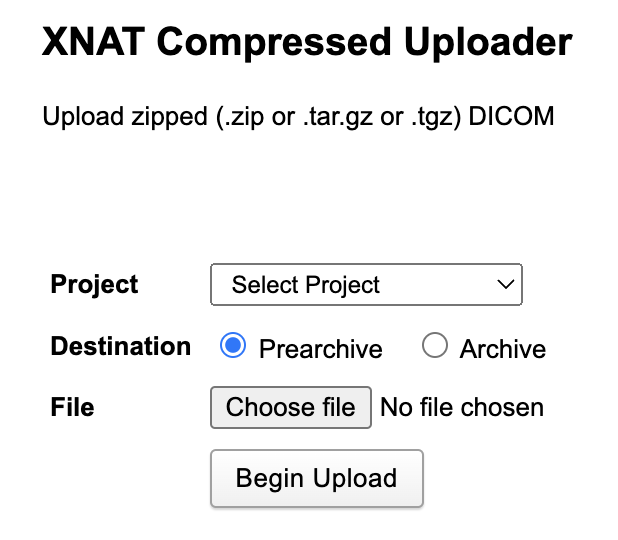
You can use XNAT’s Compressed Uploader if you are already completely confident that your files contain no patient-identifiable data. Conceptually, this is the easiest method to use and does not require anything additional to be installed. You simply interact with the same web browser you are using to access your regular XNAT session. Use the dropdown menu to select the project where the data need to end up. Choose between sending the data directly to XNAT’s archive (our usual recommendation) or the “prearchive” holding area. Then simply press the Choose file button and navigate to a single zip file into which you have previously compressed all the required data. Then click on Begin Upload. That’s all there is to it!
However, to use this method it’s vital to verify that your DICOM files are completely deidentified. This can be problematic. Metadata can be inspected using a tool like DicomBrowser (see above). You might also need to use a tool such as MicroDicom or RadiAnt (PC) or OsiriX/Horos (Mac) to verify that there are no annotations containing patient data in the image pixels themselves.
How do I make sure that only the right people have access to the data from my project?
This is something that the XNAT Team manages on your behalf. As part of the project setup process – see above – the details of all the staff who will need userids on the XNAT platform are supplied to us. We create the XNAT accounts and manage training and associated paperwork. If additional users need to be added during the lifetime of the study, then just let us know by filling in a access request form.
How can one of my collaborators get secure programmatic access to the data from my project from a remote location?
As soon as a user is given access to the XNAT web platform and can see the data within your project, the ability to access data programmatically follows automatically. XNAT has a very rich Application Programming Interface (API) that allows metadata to be queried and image data to be downloaded, as well as a whole host of other management activities. A very popular way of interacting with XNAT is via a Python program, using the XNATpy or PyXNAT libraries, but access from other languages (e.g., Java) is also possible.
How do I demonstrate to my local NHS Trust IG and InfoSec teams that XNAT meets the standards to allow me to send data to your platform.
We have a whole page on the site devoted to precisely this topic.
What documentation is there about XNAT?
The answer is lots!
- If you want to know all the detailed information about the ICR/RMH XNAT Repository, then please take a look at this document, which explains both the background to the platform and also the Standard Operating Procedures for a number of common activities.
- For general information about XNAT, visit the website of the core XNAT development team, which is now based in the company Flywheel.
- For the complete set of XNAT online documentation take a look at this index page.
How do I work with data on both sides of the NHS firewall?
This is an area that the ICR/RM XNAT Repository specialises in.
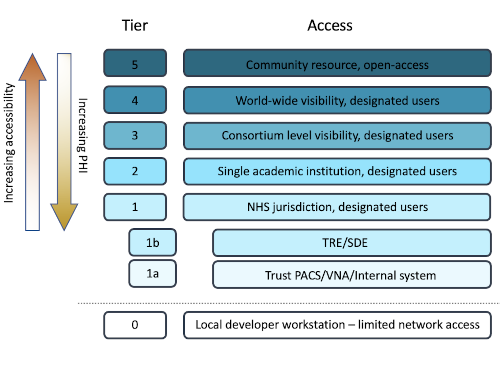
We have instances of XNAT in a variety of different contexts, which we call “data tiers”.
The lowest tier sits within a UK NHS Trust and contains identifiable data. Increasingly, there is a desire to conduct research within the NHS environment and the UK government reiterated in October 2023 the intention to implement a data access model whereby the majority of image data would remain permanently inside the NHS firewall. Even so, for research purposes, data still need to be deidentified so that patients can have confidence that researchers will not see their details. Thus, we manage an XNAT instance for pseudonymised data within an NHS Trusted Research environment.
Data for consented clinical trials may be shared outside the NHS and so additional tiers exist suitable for different types of activity.